Coronavirus
Jump to navigation
Jump to search
Coronaviruses are a group of related viruses that cause diseases in mammals and birds. In humans, coronaviruses cause respiratory tract infections that can be mild, such as some cases of the common cold (among other possible causes, predominantly rhinoviruses), and others that can be lethal, such as SARS, MERS, and COVID-19. Symptoms in other species vary: in chickens, they cause an upper respiratory tract disease, while in cows and pigs they cause diarrhea. There are yet to be vaccines or antiviral drugs to prevent or treat human coronavirus infections.
Coronaviruses constitute the subfamily Orthocoronavirinae, in the family Coronaviridae, order Nidovirales, and realm Riboviria.[5][6] They are enveloped viruses with a positive-sense single-stranded RNA genome and a nucleocapsid of helical symmetry. The genome size of coronaviruses ranges from approximately 27 to 34 kilobases, the largest among known RNA viruses.[7] The name coronavirus is derived from the Latin corona, meaning "crown" or "halo", which refers to the characteristic appearance reminiscent of a crown or a solar corona around the virions (virus particles) when viewed under two-dimensional transmission electron microscopy, due to the surface being covered in club-shaped protein spikes.
| Orthocoronavirinae | |
|---|---|
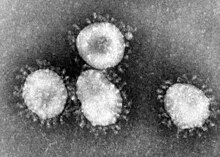
| |
| Transmission electron micrograph (TEM) of avian infectious bronchitis virus | |
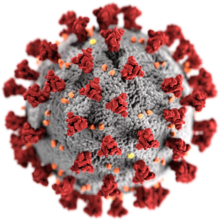
| |
| Illustration of the morphology of coronaviruses; the club-shaped viral spike peplomers, colored red, create the look of a corona surrounding the virion when observed with an electron microscope. | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Phylum: | incertae sedis |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Subfamily: | Orthocoronavirinae |
| Genera[1] | |
| Synonyms[2][3][4] | |
| |
Coronaviruses constitute the subfamily Orthocoronavirinae, in the family Coronaviridae, order Nidovirales, and realm Riboviria.[5][6] They are enveloped viruses with a positive-sense single-stranded RNA genome and a nucleocapsid of helical symmetry. The genome size of coronaviruses ranges from approximately 27 to 34 kilobases, the largest among known RNA viruses.[7] The name coronavirus is derived from the Latin corona, meaning "crown" or "halo", which refers to the characteristic appearance reminiscent of a crown or a solar corona around the virions (virus particles) when viewed under two-dimensional transmission electron microscopy, due to the surface being covered in club-shaped protein spikes.
Discovery
Human coronaviruses were first discovered in the late 1960s.[8] The earliest ones discovered were an infectious bronchitis virus in chickens and two in human patients with the common cold (later named human coronavirus 229E and human coronavirus OC43).[9] Other members of this family have since been identified, including SARS-CoV in 2003, HCoV NL63 in 2004, HKU1 in 2005, MERS-CoV in 2012, and SARS-CoV-2 (formerly known as 2019-nCoV) in 2019. Most of these have involved serious respiratory tract infections.Etymology
The name "coronavirus" is derived from Latin corona, meaning "crown" or "wreath", itself a borrowing from Greek κορώνη korṓnē, "garland, wreath". The name refers to the characteristic appearance of virions (the infective form of the virus) by electron microscopy, which have a fringe of large, bulbous surface projections creating an image reminiscent of a crown or of a solar corona. This morphology is created by the viral spike peplomers, which are proteins on the surface of the virus.[10][11]Morphology
Cross-sectional model of a coronavirus
The viral envelope consists of a lipid bilayer where the membrane (M), envelope (E) and spike (S) structural proteins are anchored.[15] A subset of coronaviruses (specifically the members of betacoronavirus subgroup A) also have a shorter spike-like surface protein called hemagglutinin esterase (HE).[5]
Inside the envelope, there is the nucleocapsid, which is formed from multiple copies of the nucleocapsid (N) protein, which are bound to the positive-sense single-stranded RNA genome in a continuous beads-on-a-string type conformation.[13][16] The lipid bilayer envelope, membrane proteins, and nucleocapsid protect the virus when it is outside the host cell.[17]
Genome
Schematic representation of the genome organization and functional domains of S protein for SARS-CoV and MERS-CoV.
The genome organization for a coronavirus is 5′-leader-UTR-replicase/transcriptase-spike (S)-envelope (E)-membrane (M)-nucleocapsid (N)-3′UTR-poly (A) tail. The open reading frames 1a and 1b, which occupy the first two-thirds of the genome, encode the replicase/transcriptase polyprotein. The replicase/transcriptase polyprotein self cleaves to form the nonstructural proteins (nsps).[19]
The later reading frames encode the four major structural proteins: spike, envelope, membrane, and nucleocapsid.[20] Interspersed between these reading frames are the reading frames for the accessory proteins. The number of accessory proteins and their function is unique depending on the specific coronavirus.[19]
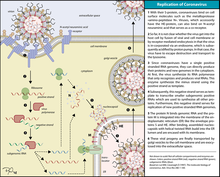
Life cycle
Entry
The life cycle of a coronavirus
On entry into the host cell, the virus particle is uncoated, and its genome enters the cell cytoplasm.[22] The coronavirus RNA genome has a 5′ methylated cap and a 3′ polyadenylated tail, which allows the RNA to attach to the host cell's ribosome for translation.[23] The host ribosome translates the initial overlapping open reading frame of the virus genome and forms a long polyprotein. The polyprotein has its own proteases which cleave the polyprotein into multiple nonstructural proteins.[19]
Replication
A number of the nonstructural proteins coalesce to form a multi-protein replicase-transcriptase complex (RTC). The main replicase-transcriptase protein is the RNA-dependent RNA polymerase (RdRp). It is directly involved in the replication and transcription of RNA from an RNA strand. The other nonstructural proteins in the complex assist in the replication and transcription process. The exoribonuclease non-structural protein, for instance, provides extra fidelity to replication by providing a proofreading function which the RNA-dependent RNA polymerase lacks.[24]One of the main functions of the complex is to replicate the viral genome. RdRp directly mediates the synthesis of negative-sense genomic RNA from the positive-sense genomic RNA. This is followed by the replication of positive-sense genomic RNA from the negative-sense genomic RNA.[19] The other important function of the complex is to transcribe the viral genome. RdRp directly mediates the synthesis of negative-sense subgenomic RNA molecules from the positive-sense genomic RNA. This is followed by the transcription of these negative-sense subgenomic RNA molecules to their corresponding positive-sense mRNAs.[19]
Release
The replicated positive-sense genomic RNA becomes the genome of the progeny viruses. The mRNAs are gene transcripts of the last third of the virus genome after the initial overlapping reading frame. These mRNAs are translated by the host's ribosomes into the structural proteins and a number of accessory proteins.[19] RNA translation occurs inside the endoplasmic reticulum. The viral structural proteins S, E, and M move along the secretory pathway into the Golgi intermediate compartment. There, the M proteins direct most protein-protein interactions required for assembly of viruses following its binding to the nucleocapsid.[25] Progeny viruses are then released from the host cell by exocytosis through secretory vesicles.[25]Transmission
Human to human transmission of coronaviruses is primarily thought to occur among close contacts via respiratory droplets generated by sneezing and coughing.[26] The interaction of the coronavirus spike protein with its complement host cell receptor is central in determining the tissue tropism, infectivity, and species range of the virus.[27][28] The SARS coronavirus, for example, infects human cells by attaching to the angiotensin-converting enzyme 2 (ACE2) receptor.[29]Taxonomy
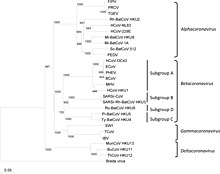
Phylogenetic tree of coronaviruses
- Genus: Alphacoronavirus
- Genus Betacoronavirus; type species: Murine coronavirus
- Species: Betacoronavirus 1, Human coronavirus HKU1, Murine coronavirus, Pipistrellus bat coronavirus HKU5, Rousettus bat coronavirus HKU9, Severe acute respiratory syndrome-related coronavirus, Tylonycteris bat coronavirus HKU4, Middle East respiratory syndrome-related coronavirus, Human coronavirus OC43, Hedgehog coronavirus 1 (EriCoV)
- Genus Gammacoronavirus; type species: Infectious bronchitis virus
- Genus Deltacoronavirus; type species: Bulbul coronavirus HKU11
Evolution
The most recent common ancestor (MRCA) of all coronaviruses has been estimated to have existed as recently as 8000 BCE, though some models place the MRCA as far back as 55 million years or more, implying long term coevolution with bats.[30] The MRCAs of the alphacoronavirus line has been placed at about 2400 BCE, the betacoronavirus line at 3300 BCE, the gammacoronavirus line at 2800 BCE, and the deltacoronavirus line at about 3000 BCE. It appears that bats and birds, as warm-blooded flying vertebrates, are ideal hosts for the coronavirus gene source (with bats for alphacoronavirus and betacoronavirus, and birds for gammacoronavirus and deltacoronavirus) to fuel coronavirus evolution and dissemination.[31]Bovine coronavirus and canine respiratory coronaviruses diverged from a common ancestor recently (~ 1950).[32] Bovine coronavirus and human coronavirus OC43 diverged around the 1890s. Bovine coronavirus diverged from the equine coronavirus species at the end of the 18th century.[33]
The MRCA of human coronavirus OC43 has been dated to the 1950s.[34]
MERS-CoV, although related to several bat coronavirus species, appears to have diverged from these several centuries ago.[35] The human coronavirus NL63 and a bat coronavirus shared an MRCA 563–822 years ago.[36]
The most closely related bat coronavirus and SARS-CoV diverged in 1986.[37] A path of evolution of the SARS virus and keen relationship with bats have been proposed. The authors suggest that the coronaviruses have been coevolved with bats for a long time and the ancestors of SARS-CoV first infected the species of the genus Hipposideridae, subsequently spread to species of the Rhinolophidae and then to civets, and finally to humans.[38][39]
Alpaca coronavirus and human coronavirus 229E diverged before 1960.[40]
Illustration of SARSr-CoV virion
Seven strains of human coronaviruses are known, of which four produce the generally mild symptoms of the common cold:
- Human coronavirus OC43 (HCoV-OC43), Betacoronavirus
- Human coronavirus HKU1, Betacoronavirus, its genome has 75% similarity to OC43[43]
- Human coronavirus 229E (HCoV-229E), Alphacoronavirus
- Human coronavirus NL63 (HCoV-NL63, New Haven coronavirus), Alphacoronavirus
- Middle East respiratory syndrome-related coronavirus (MERS-CoV), previously known as novel coronavirus 2012 and HCoV-EMC
- Severe acute respiratory syndrome coronavirus (SARS-CoV or "SARS-classic")
- Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), previously known as 2019-nCoV or "novel coronavirus 2019"
Outbreaks of coronavirus types of relatively high mortality are as follows:
Severe acute respiratory syndrome (SARS)
| SARS-CoV-2 | MERS-CoV | SARS-CoV | |
|---|---|---|---|
| Demographic | |||
| Date of first identified case |
December 2019 |
June 2012 |
November 2002 |
| Location of first identified case |
Wuhan, China |
Jeddah, Saudi Arabia |
Shunde, China |
| Age average | 56[51] | 56 | 39.9 |
| Age range | all ages | 14–94 | 1–91 |
| Male:female ratio | 1.6:1[51] | 3.3:1 | 1:1.25 |
| Confirmed cases | 713,171[49][a] | 2494 | 8096 |
| Deaths | 33,597[49][a] | 858 | 744 |
| Case fatality rate | 3.4% | 37% | 10% |
| Health-care workers | 8%[52] | 9.8% | 23.1% |
| Symptoms | |||
| Fever | 87.9%[53] | 98% | 99–100% |
| Dry cough | 67.7%[53] | 47% | 29–75% |
| Dyspnea | 18.6%[53] | 72% | 40–42% |
| Diarrhea | 3.7%[53] | 26% | 20–25% |
| Sore throat | 13.9%[53] | 21% | 13–25% |
| Ventilatory support | 4.1%[54] | 24.5%[55] | 14–20% |
| Notes | |||
|
Middle East respiratory syndrome (MERS)
In September 2012, a new type of coronavirus was identified, initially called Novel Coronavirus 2012, and now officially named Middle East respiratory syndrome coronavirus (MERS-CoV).[56][57] The World Health Organization issued a global alert soon after.[58] The WHO update on 28 September 2012 said the virus did not seem to pass easily from person to person.[59] However, on 12 May 2013, a case of human-to-human transmission in France was confirmed by the French Ministry of Social Affairs and Health.[60] In addition, cases of human-to-human transmission were reported by the Ministry of Health in Tunisia. Two confirmed cases involved people who seemed to have caught the disease from their late father, who became ill after a visit to Qatar and Saudi Arabia. Despite this, it appears the virus had trouble spreading from human to human, as most individuals who are infected do not transmit the virus.[61] By 30 October 2013, there were 124 cases and 52 deaths in Saudi Arabia.[62]After the Dutch Erasmus Medical Centre sequenced the virus, the virus was given a new name, Human Coronavirus–Erasmus Medical Centre (HCoV-EMC). The final name for the virus is Middle East respiratory syndrome coronavirus (MERS-CoV). In May 2014, the only two United States cases of MERS-CoV infection were recorded, both occurring in healthcare workers who worked in Saudi Arabia and then travelled to the U.S. One was treated in Indiana and one in Florida. Both were hospitalized temporarily and then discharged.[63]
In May 2015, an outbreak of MERS-CoV occurred in the Republic of Korea, when a man who had traveled to the Middle East, visited 4 hospitals in the Seoul area to treat his illness. This caused one of the largest outbreaks of MERS-CoV outside the Middle East.[64] As of December 2019, 2,468 cases of MERS-CoV infection had been confirmed by laboratory tests, 851 of which were fatal, a mortality rate of approximately 34.5%.[65]
In December 2019, a pneumonia outbreak was reported in Wuhan, China.[66] On 31 December 2019, the outbreak was traced to a novel strain of coronavirus,[67] which was given the interim name 2019-nCoV by the World Health Organization (WHO),[68][69][70] later renamed SARS-CoV-2 by the International Committee on Taxonomy of Viruses. Some researchers have suggested that the Huanan Seafood Wholesale Market may not be the original source of viral transmission to humans.[71][72]
As of 29 March 2020, there have been at least 33,597[49] confirmed deaths and more than 713,171[49] confirmed cases in the coronavirus pneumonia pandemic. The Wuhan strain has been identified as a new strain of Betacoronavirus from group 2B with approximately 70% genetic similarity to the SARS-CoV.[73] The virus has a 96% similarity to a bat coronavirus, so it is widely suspected to originate from bats as well.[71][74] The pandemic has resulted in travel restrictions and nationwide lockdowns in several countries.
Other animals
Coronaviruses have been recognized as causing pathological conditions in veterinary medicine since the early 1970s. Except for avian infectious bronchitis, the major related diseases have mainly an intestinal location.[75]Diseases caused
Coronaviruses primarily infect the upper respiratory and gastrointestinal tract of mammals and birds. They also cause a range of diseases in farm animals and domesticated pets, some of which can be serious and are a threat to the farming industry. In chickens, the infectious bronchitis virus (IBV), a coronavirus, targets not only the respiratory tract but also the urogenital tract. The virus can spread to different organs throughout the chicken.[76] Economically significant coronaviruses of farm animals include porcine coronavirus (transmissible gastroenteritis coronavirus, TGE) and bovine coronavirus, which both result in diarrhea in young animals. Feline coronavirus: two forms, feline enteric coronavirus is a pathogen of minor clinical significance, but spontaneous mutation of this virus can result in feline infectious peritonitis (FIP), a disease associated with high mortality. Similarly, there are two types of coronavirus that infect ferrets: Ferret enteric coronavirus causes a gastrointestinal syndrome known as epizootic catarrhal enteritis (ECE), and a more lethal systemic version of the virus (like FIP in cats) known as ferret systemic coronavirus (FSC).[77] There are two types of canine coronavirus (CCoV), one that causes mild gastrointestinal disease and one that has been found to cause respiratory disease. Mouse hepatitis virus (MHV) is a coronavirus that causes an epidemic murine illness with high mortality, especially among colonies of laboratory mice.[78] Sialodacryoadenitis virus (SDAV) is highly infectious coronavirus of laboratory rats, which can be transmitted between individuals by direct contact and indirectly by aerosol. Acute infections have high morbidity and tropism for the salivary, lachrymal and harderian glands.[79]A HKU2-related bat coronavirus called swine acute diarrhea syndrome coronavirus (SADS-CoV) causes diarrhea in pigs.[80]
Prior to the discovery of SARS-CoV, MHV had been the best-studied coronavirus both in vivo and in vitro as well as at the molecular level. Some strains of MHV cause a progressive demyelinating encephalitis in mice which has been used as a murine model for multiple sclerosis. Significant research efforts have been focused on elucidating the viral pathogenesis of these animal coronaviruses, especially by virologists interested in veterinary and zoonotic diseases.[81]
In domestic animals
- Infectious bronchitis virus (IBV) causes avian infectious bronchitis.
- Porcine coronavirus (transmissible gastroenteritis coronavirus of pigs, TGEV).[82][83]
- Bovine coronavirus (BCV), responsible for severe profuse enteritis in of young calves.
- Feline coronavirus (FCoV) causes mild enteritis in cats as well as severe Feline infectious peritonitis (other variants of the same virus).
- the two types of canine coronavirus (CCoV) (one causing enteritis, the other found in respiratory diseases).
- Turkey coronavirus (TCV) causes enteritis in turkeys.
- Ferret enteric coronavirus causes epizootic catarrhal enteritis in ferrets.
- Ferret systemic coronavirus causes FIP-like systemic syndrome in ferrets.[84]
- Pantropic canine coronavirus.
- Rabbit enteric coronavirus causes acute gastrointestinal disease and diarrhea in young European rabbits. Mortality rates are high.[85]
- Porcine epidemic diarrhea virus (PED or PEDV), has emerged around the world.[86]
Genomic cis-acting elements
In common with the genomes of all other RNA viruses, coronavirus genomes contain cis-acting RNA elements that ensure the specific replication of viral RNA by a virally encoded RNA-dependent RNA polymerase. The embedded cis-acting elements devoted to coronavirus replication constitute a small fraction of the total genome, but this is presumed to be a reflection of the fact that coronaviruses have the largest genomes of all RNA viruses. The boundaries of cis-acting elements essential to replication are fairly well-defined, and the RNA secondary structures of these regions are understood. However, how these cis-acting structures and sequences interact with the viral replicase and host cell components to allow RNA synthesis is not well understood.[87][5]Genome packaging
The assembly of infectious coronavirus particles requires the selection of viral genomic RNA from a cellular pool that contains an abundant excess of non-viral and viral RNAs. Among the seven to ten specific viral mRNAs synthesized in virus-infected cells, only the full-length genomic RNA is packaged efficiently into coronavirus particles. Studies have revealed cis-acting elements and trans-acting viral factors involved in the coronavirus genome encapsidation and packaging. Understanding the molecular mechanisms of genome selection and packaging is critical for developing antiviral strategies and viral expression vectors based on the coronavirus genome.[87][5]See also
References
CoVs also have the largest known RNA virus genomes, ranging from 27 to 34 kb (31, 32), and increased fidelity in CoVs is likely required for the maintenance of these large genomes (14).
[T]here is also a characteristic "fringe" of projections 200 A long, which are rounded or petal shaped ... This appearance, recalling the solar corona, is shared by mouse hepatitis virus and several viruses recently recovered from man, namely strain B814, 229E and several others.
[T]hese viruses displayed a characteristic fringe of large, distinctive, petal-shaped peplomers or spikes which resembled a crown, like the corona spinarum in religious art; hence the name coronaviruses.
Virions acquired an envelope by budding into the cisternae and formed mostly spherical, sometimes pleomorphic, particles that averaged 78 nm in diameter (Figure 1A).
See section: Virion Structure.
Particle diameters ranged from 50 to 150 nm, excluding the spikes, with mean particle diameters of 82 to 94 nm; Also See Figure 1 for double shell.
See Figure 4c.
See Figure 10.
See Figure 1.
See Figure 2.
Finally, these results, combined with those from previous work (33, 44), suggest that CoVs encode at least three proteins involved in fidelity (nsp12-RdRp, nsp14-ExoN, and nsp10), supporting the assembly of a multiprotein replicase-fidelity complex, as described previously (38).
See section: Coronavirus Life Cycle—Assembly and Release
Nevertheless, the interaction between S protein and receptor remains the principal, if not sole, determinant of coronavirus host species range and tissue tropism.
Different SARS-CoV strains isolated from several hosts vary in their binding affinities for human ACE2 and consequently in their infectivity of human cells76,78 (Fig. 6b)
- Thiel V (editor). (2007). Coronaviruses: Molecular and Cellular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-16-5.[page needed]
Further reading
| Wikimedia Commons has media related to Coronaviridae. |
| Wikispecies has information related to Orthocoronavirinae |
| Look up coronavirus in Wiktionary, the free dictionary. |
- Alwan A, Mahjour J, Memish ZA (2013). "Novel coronavirus infection: time to stay ahead of the curve". Eastern Mediterranean Health Journal. 19 Suppl 1: S3-4. doi:10.26719/2013.19.supp1.S3. PMID 23888787.
- Laude H, Rasschaert D, Delmas B, Godet M, Gelfi J, Charley B (June 1990). "Molecular biology of transmissible gastroenteritis virus". Veterinary Microbiology. 23 (1–4): 147–54. doi:10.1016/0378-1135(90)90144-K. PMID 2169670.
- Sola I, Alonso S, Zúñiga S, Balasch M, Plana-Durán J, Enjuanes L (April 2003). "Engineering the transmissible gastroenteritis virus genome as an expression vector inducing lactogenic immunity". Journal of Virology. 77 (7): 4357–69. doi:10.1128/JVI.77.7.4357-4369.2003. PMC 150661. PMID 12634392.
- Tajima M (1970). "Morphology of transmissible gastroenteritis virus of pigs. A possible member of coronaviruses. Brief report". Archiv für die Gesamte Virusforschung. 29 (1): 105–8. doi:10.1007/BF01253886. PMID 4195092.
| Classification |
|---|



কোন মন্তব্য নেই:
একটি মন্তব্য পোস্ট করুন